Abstract
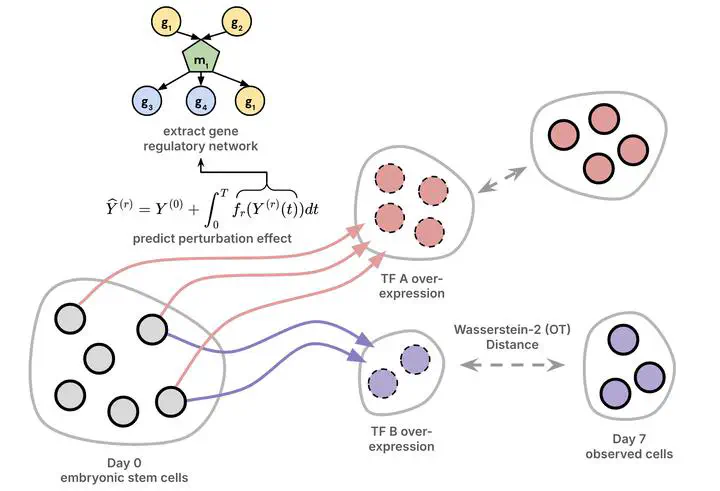
Modern high-throughput biological datasets with thousands of perturbations provide the opportunity for large-scale discovery of causal graphs that represent the regulatory interactions between genes. Differentiable causal graphical models have been proposed to infer a gene regulatory network (GRN) from large scale interventional datasets, capturing the causal gene regulatory relationships from genetic perturbations. However, existing models are limited in their expressivity and scalability while failing to address the dynamic nature of biological processes such as cellular differentiation. We propose PerturbODE, a novel framework that incorporates biologically informative neural ordinary differential equations (neural ODEs) to model cell state trajectories under perturbations and derive the causal GRN from the neural ODE’s parameters. We demonstrate PerturbODE’s efficacy in trajectory prediction and GRN inference across simulated and real over-expression datasets.